ATAC Sequencing
Assays for Transposase-Accessible Chromatin using sequencing, short for ATAC-Seq, is a next-generation sequencing (NGS)-based method to study the open regions of chromatin on a genome-wide scale. ATAC-Seq has the advantages of easy operation, no crosslinking required, high signal-to-noise ratio, and low requirement for sample amount. Combined the data of ATAC-Seq, RNA-Seq, and epigenomics, the relationship between gene differential expression and regulation can be studied more accurately and comprehensively.
Overview
The DNA of eukaryotes is tightly bound to a group of proteins called histones, which constitute half the mass of a eukaryotic chromosome. There are numerous units that DNA enwinds histone formed a bead like structure called nucleosome, and the chromatin looks like many beads on a string. DNA replication and gene transcription require the unwinding of the higher structure of DNA. Open chromatin regions are these folded regions that contain important binding sites for transcription factors and transcriptional regulatory elements. ATAC-Seq utilizes the hyperactive mutant Tn5 transposase to insert sequencing adapters into accessible DNA regions, through which the open chromatin sequence can be separated from the entire chromatin, followed by next-generation sequencing and corresponding bioinformatics analysis. The research on open chromatin addresses the needs for deeper understanding of gene regulation, transcription factors that relate to cell physiology and diseases, etc.
Features
| High Throughput | Genome-Wide | Bioinformatics Analysis | One-stop Service |
|---|---|---|---|
| Cost-effective transcriptional profiling solution for large sample-size assays. | Profile all open chromatins, known and novel. | Our integrated bioinformatics pipeline can be tailored to suit your project. | Provides one-stop service for library construction, sequencing, sample QC and data analysis. |
Project Workflow

1. Sample Preparation
DNA purification;
quality assessment and quantification.

2. Library Preparation
DNA fragmentation;
library preparation.

3. Sequencing
Illumina HiSeq;
PE 50/75/100/150.

4. Data Analysis
Visualize and preprocess results, and perform custom bioinformatics analysis.
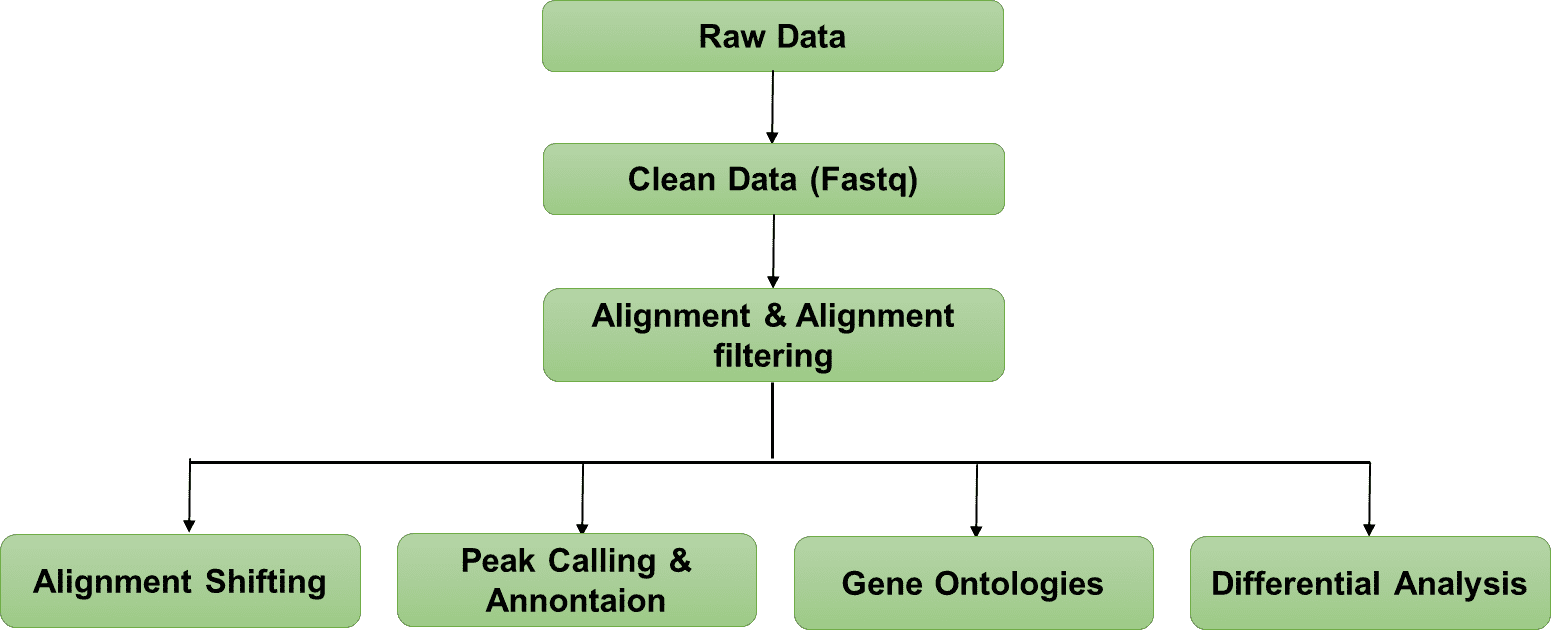
Bioinformatics Analysis Pipeline
In-depth data analysis:
- Data quality control;
- Peak calling and annotation;
- Fragment size distribution;
- Gene ontologies;
- KEGG pathways;
- Differential analysis.
Sample Requirements
Cell sample ≥ 50,000 cells; Nucleus sample ≥ 50,000 nuclei
Sample storage: Nuclei can be frozen at –20˚C for up to two days, avoid repeated freezing and thawing.
Shipping Method: Samples are supposed to stored in a 1.5 mL microcentrifuge tube, sealed with sealing film. Shipments are generally recommended to contain 5-10 pounds of dry ice per 24 hours.
Deliverable: FastQ, BAM, coverage summary, QC report, full statistical analysis & alignments, custom analysis reports on customer request.
References:
- Satpathy AT, Saligrama N, Buenrostro JD, et al. Transcript-indexed ATAC-seq for precision immune profiling. Nature medicine. 2018;24(5):580-90.
- Divate M, Cheung E. GUAVA: a Graphical User Interface for the Analysis and Visualization of ATAC-seq Data. Frontiers in genetics. 2018;9:250.
- Corces MR, Trevino AE, Hamilton EG, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nature methods. 2017;14(10):959-62.
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC‐seq: a method for assaying chromatin accessibility genome‐wide. Curr Protoc Mol Biol. 2015;109(1):21.9. 1-.9. 9.


