LncRNA and CircRNA RIP Sequencing
Our service is designed to provide researchers with a comprehensive understanding of long non-coding RNAs (LncRNAs) and circular RNAs (CircRNAs) and their interactions with RNA-binding proteins, based on our RIP Sequencing platform.
Overview of LncRNAs and CircRNAs
LncRNAs and CircRNAs are two fascinating classes of non-coding RNAs that have gained immense attention due to their regulatory roles in gene expression and cellular processes.
LncRNAs, although not encoding proteins, wield remarkable influence over cellular function. They are lengthy RNA molecules that orchestrate a symphony of gene regulatory activities. By interacting with chromatin-modifying enzymes and recruiting them to specific genomic regions, LncRNAs sculpt the landscape of DNA and histone modifications. This dynamic choreography affects chromatin structure, accessibility, and ultimately gene expression. As molecular architects, LncRNAs guide the cell in adapting genomic functions to developmental cues and environmental signals, ushering genes into appropriate expression patterns.
CircRNAs are marked by their unique closed-loop structure, distinct from the linear form of most RNA molecules. This distinctive topology enables them to engage in intricate intracellular regulatory networks. While the full spectrum of CircRNA functions is still being uncovered, it's evident that they serve as essential participants in fine-tuning cellular processes. Their potential roles in sponging microRNAs, interacting with RNA-binding proteins, and influencing translation make them intriguing candidates for a wide range of regulatory tasks.
RNA Immunoprecipitation (RIP) sequencing is a powerful technique that allows researchers to investigate the interactions between RNA molecules and RNA-binding proteins (RBPs). By isolating the complexes formed between LncRNAs/CircRNAs and RBPs, we can decipher the regulatory roles of these non-coding RNAs and shed light on the intricate mechanisms governing gene expression.
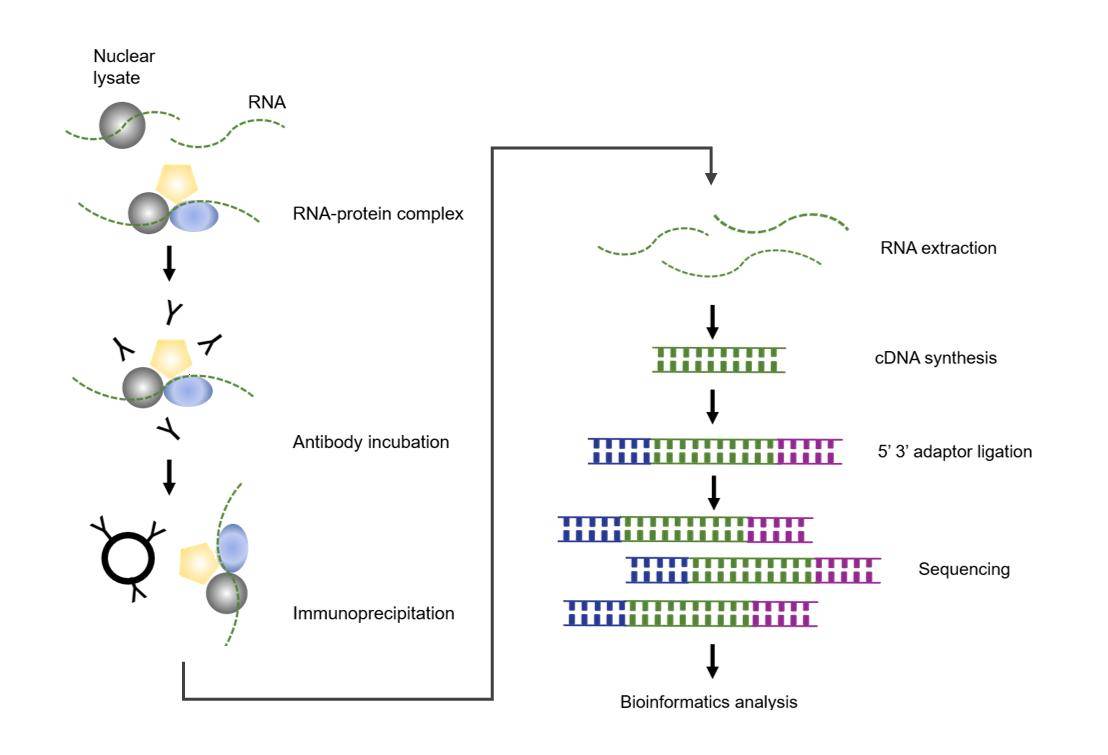 Workflow of RIP Sequencing – CD Genomics
Workflow of RIP Sequencing – CD Genomics
Features
| High Resolution |
High Coverage |
Cutting-edge Algorithms |
All-in-One Convenience |
| Enable the identification of even the most delicate protein binding sites across the entire genome. | Capture intricate details and reveal the subtleties of RNA interactions that might otherwise remain hidden. | Enable you to explore interactions, pathways, and networks across multiple layers, uncovering the interplay between RNAs and proteins. | One-stop solution, covering everything from library construction and sequencing to sample quality control and data analysis. |
Our LncRNA and CircRNA RIP Sequencing Workflow
- Sample Preparation: We work with a variety of sample types, including cell lines and tissues. Our optimized protocols ensure efficient enrichment of LncRNAs and CircRNAs bound to RNA-binding proteins (RBPs).
- Immunoprecipitation (RIP): Our RIP protocol enables the isolation of RNA molecules that interact with specific RBPs, providing insights into the regulatory networks involving LncRNAs and CircRNAs.
- Library Preparation: We generate high-quality RNA libraries suitable for next-generation sequencing, ensuring accurate and reproducible results.
- Sequencing: Utilizing cutting-edge sequencing platforms, we generate deep sequencing data to capture a comprehensive snapshot of the LncRNA and CircRNA expression landscape.
- Bioinformatics Analysis: Our bioinformatics experts process and analyze the sequencing data, employing advanced algorithms and tools to identify differentially expressed LncRNAs and CircRNAs, predict potential interactions, and uncover underlying biological mechanisms.
- Data Interpretation: We provide detailed reports summarizing the analysis results, including visualizations and annotations, to help you interpret the findings in the context of your research.
Case Studies
-
-
circPTPRA in Bladder Cancer Progression Through IGF2BP1 Regulation
-
Background
Bladder cancer (BC) continues to be a significant global health concern, prompting extensive research efforts to uncover its underlying molecular mechanisms and potential therapeutic targets. One key player in the context of BC is the insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), a critical reader of N6-methyladenosine (m6A) modifications. This case study focuses on a comprehensive investigation into the interplay between circRNAs, IGF2BP1, and their impact on BC progression.
Methods
The research team embarked on a multifaceted study to unravel the role of circRNAs in BC, particularly their interactions with IGF2BP1.
- Clinical Role of IGF2BP1 in BC: Initial investigations revolved around understanding the clinical relevance of IGF2BP1 in BC. This involved an analysis of patient samples to gauge the expression levels of IGF2BP1 and its potential correlation with disease progression.
- RIP-seq Analysis for circRNA Identification: RNA immunoprecipitation sequencing (RIP-seq) was employed to identify circRNAs that interact with IGF2BP1 in BC cells. This step aimed to pinpoint specific circRNAs that might have regulatory roles through interaction with IGF2BP1.
- Functional Studies in BC Cell Lines and Animal Models: Candidate circRNA circPTPRA was subjected to functional investigations using BC cell lines and animal xenograft studies. These studies aimed to elucidate the impact of circPTPRA on key cellular processes such as proliferation, migration, and invasion.
- Regulation Effects of circPTPRA on IGF2BP1: The team evaluated how circPTPRA influenced the regulatory activity of IGF2BP1. The downstream effects of circPTPRA on IGF2BP1-regulated genes were studied through techniques such as RNA sequencing.
- Molecular Mechanisms of circPTPRA: To gain insights into the molecular mechanisms at play, the researchers probed into how circPTPRA functioned as a blocker in the recognition of m6A-modified RNAs. This involved investigating the interaction between circPTPRA and the KH domains of IGF2BP1.
Results
The study yielded several significant findings:
- The binding of IGF2BP1 to circPTPRA was predominant in the cytoplasm of BC cells.
 Overlap of IGF2BP1-binding circRNAs identified by RIP–seq. (Xie et al., 2021)
Overlap of IGF2BP1-binding circRNAs identified by RIP–seq. (Xie et al., 2021)
- Overexpression of circPTPRA counteracted the pro-tumorigenic effects induced by IGF2BP1, including cell proliferation, migration, and invasion.
- CircPTPRA exerted its effects by downregulating IGF2BP1-mediated regulation of MYC and FSCN1 expression.
 CircPTPRA downregulated MYC and FSCN1 expression via interacting with IGF2BP1 in BC cells. (Xie et al., 2021)
CircPTPRA downregulated MYC and FSCN1 expression via interacting with IGF2BP1 in BC cells. (Xie et al., 2021)
- Importantly, the interaction of circPTPRA with the KH domains of IGF2BP1 interfered with the recognition of m6A-modified RNAs.
-
-
LINC00941 - A Novel Therapeutic Target for IPF via Autophagy Regulation
-
Background
Idiopathic Pulmonary Fibrosis (IPF) is a devastating lung disease characterized by progressive scarring of lung tissue, leading to compromised respiratory function and reduced quality of life. Despite extensive research, effective treatments for IPF remain elusive. The hallmark features of IPF include fibroblast-to-myofibroblast differentiation, myofibroblast proliferation, and migration, all of which contribute to the pathogenesis of the disease. Addressing these processes presents a viable therapeutic strategy. Recent investigations have identified LINC00941 (also known as lncIAPF) as a potentially critical player in driving pulmonary fibrosis by promoting these key pathological events.
Methods
The study aimed to elucidate the mechanisms by which LINC00941 contributes to the progression of IPF. A multidisciplinary approach involving various experimental techniques was employed:
- Promoter Activation Analysis: Researchers used Assay for Transposase-Accessible Chromatin using Sequencing (ATAC-seq) and Chromatin Immunoprecipitation (ChIP) experiments to identify histone 3 lysine 27 acetylation (H3K27ac) as a key epigenetic modification that activated the LINC00941 promoter.
- Transcription Factor Interaction Study: Using ChIP and subsequent sequencing (ChIP-seq), the study determined that the transcription factor ATF3 binds to the activated region on the LINC00941 promoter, leading to enhanced transcription.
- RNA-Protein Complex Characterization: RNA affinity isolation, RNA immunoprecipitation (RIP), RNase-RIP, half-life analysis, and ubiquitination assays were employed to unveil the formation of an RNA-protein complex between LINC00941 and ELAVL1/HuR.
- Target Gene Regulation Analysis: ELAVL1 RIP-seq, RIP-PCR, mRNA stability, and rescue experiments unveiled the impact of the LINC00941-ELAVL1 complex on the stability of target genes, including EZH2, STAT1, and FOXK1.
Results
The study's findings shed light on the intricate molecular mechanisms underlying the contribution of LINC00941 to IPF:
- Activation of LINC00941: H3K27ac modification activated the LINC00941 promoter, enhancing its transcription.
- ATF3 Binding: ATF3, a transcription factor, bound to the activated region on the LINC00941 promoter, facilitating increased transcription.
- RNA-Protein Complex: LINC00941 formed a complex with ELAVL1/HuR, a critical step for its pro-fibrotic effects.
- Autophagy Inhibition: The LINC00941-ELAVL1 axis hindered autophagosome-lysosome fusion, leading to autophagy inhibition.
- Target Gene Regulation: The complex controlled the stability of specific target genes, influencing the autophagy process.
 Highly expressed mechanism of LINC00941/lncIAPF. (Zhang et al., 2022)
Highly expressed mechanism of LINC00941/lncIAPF. (Zhang et al., 2022)
Sample Requirements
For sample volumes ≥ 200 ng: Our standard library preparation procedures will be employed.
For sample volumes ≤ 10 ng: Our library preparation approach will be utilized to accommodate lower input amounts without compromising data quality.
Tissue Samples: A minimum of ≥ 100 mg of tissue is required for optimal results.
Cell Samples: Cell samples must not be fewer than 107 cells.
Mycoplasma Contamination: Cell samples must be free of mycoplasma contamination to ensure reliable results.
Sample Storage: RNA can be dissolved in ethanol or RNA-free ultra-pure water and stored at -80°C. RNA should avoid repeated freezing and thawing.
Shipping Method: When shipping RNA samples, the RNA sample is stored in a 1.5 mL Eppendorf tube, sealed with sealing film. Shipments are generally recommended to contain 5-10 pounds of dry ice per 24 hours.
Deliverable: Comprehensive RIP experiment report. Fastq, BAM, coverage summary, QC report, custom bioinformatics analysis.
References:
- Xie, Fei, et al. "CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression." Molecular cancer 20.1 (2021): 1-17.
- Zhang, Jinjin, et al. "ATF3-activated accelerating effect of LINC00941/lncIAPF on fibroblast-to-myofibroblast differentiation by blocking autophagy depending on ELAVL1/HuR in pulmonary fibrosis." Autophagy 18.11 (2022): 2636-2655.
* For Research Use Only. Not for use in diagnostic procedures.
 Workflow of RIP Sequencing – CD Genomics
Workflow of RIP Sequencing – CD Genomics  Overlap of IGF2BP1-binding circRNAs identified by RIP–seq. (Xie et al., 2021)
Overlap of IGF2BP1-binding circRNAs identified by RIP–seq. (Xie et al., 2021)  CircPTPRA downregulated MYC and FSCN1 expression via interacting with IGF2BP1 in BC cells. (Xie et al., 2021)
CircPTPRA downregulated MYC and FSCN1 expression via interacting with IGF2BP1 in BC cells. (Xie et al., 2021)  Highly expressed mechanism of LINC00941/lncIAPF. (Zhang et al., 2022)
Highly expressed mechanism of LINC00941/lncIAPF. (Zhang et al., 2022) 
