piRNA Sequencing
Piwi-interacting RNA sequencing (piRNA-seq) is a next-generation sequencing (NGS)-based method to comprehensively capture, sequence and analyze piRNA sequences (26-32nt) in animal cells. These small fragments function primarily by interacting with PIWI (P-element-induced wimpy testis) family proteins to form RNA-protein complexes. Our piRNA-seq service is dedicated to detecting and analyzing all piRNAs in a sample, providing a powerful tool for piRNA screening and research.
Overview
The piRNAs are the largest class of single-stranded, small non-coding RNAs of about 26-32 nucleotides. Two types of piRNAs have been identified based on their genomic origins: repetitive sequence-derived piRNAs and non-repetitive sequence-derived piRNAs. The piRNAs are commonly expressed only in totipotent animal cells, such as germ cells, tumor cells and stem cells in a tissue-specific manner. piRNA performs biological functions by combining with PIWI subfamily proteins, which are mainly expressed in the germline, to form piRNA complexes (piRC) and are involved in epigenetic, transcriptional and post-transcriptional silencing of transposons and other genetic elements, maintaining the functions of germ cells and stem cells, and regulating mRNA translation and stability. In addition, evidences are arising that piRNA is abnormally expressed in multiple cancers, which suggest piRNAs may be involved in the occurence and development of tumors. piRNA sequencing (piRNA-seq) is a sequencing technology specifically designed to capture piRNA from animal cells, allowing the detection of piRNAs isoforms and novel piRNAs. piRNA-seq provides insight into transposon silencing, germline development, spermatogenesis, and how they maintain genomic integrity. In clinical research, especially cancer research, piRNA-seq can be used for elucidating mechanisms underlying diseases, biomarker screening, and development of targeted therapies.
Features
| Application | High Efficiency | Low Input | One-Stop Solution |
|---|---|---|---|
| This method can profile and quantify known piRNAs and new piRNAs. | Deliver millions of sequences and detect multiple copies of piRNA at a time. | Highly specialized library preparation method, allowing low input RNA of about 2 ug. | One-stop solution from sample preparation, library construction, to sequencing and data analysis. |
Project Workflow

1. Sample Preparation
Quality assessment and quantification; RNA purification.

2. Library Preparation
Size selection (24-35nt); strand-specific libraries.

3. Sequencing
Illumina Illumina HiSeq;
PE50.

4. Data Analysis
Visualize and preprocess results, and perform custom bioinformatics analysis.
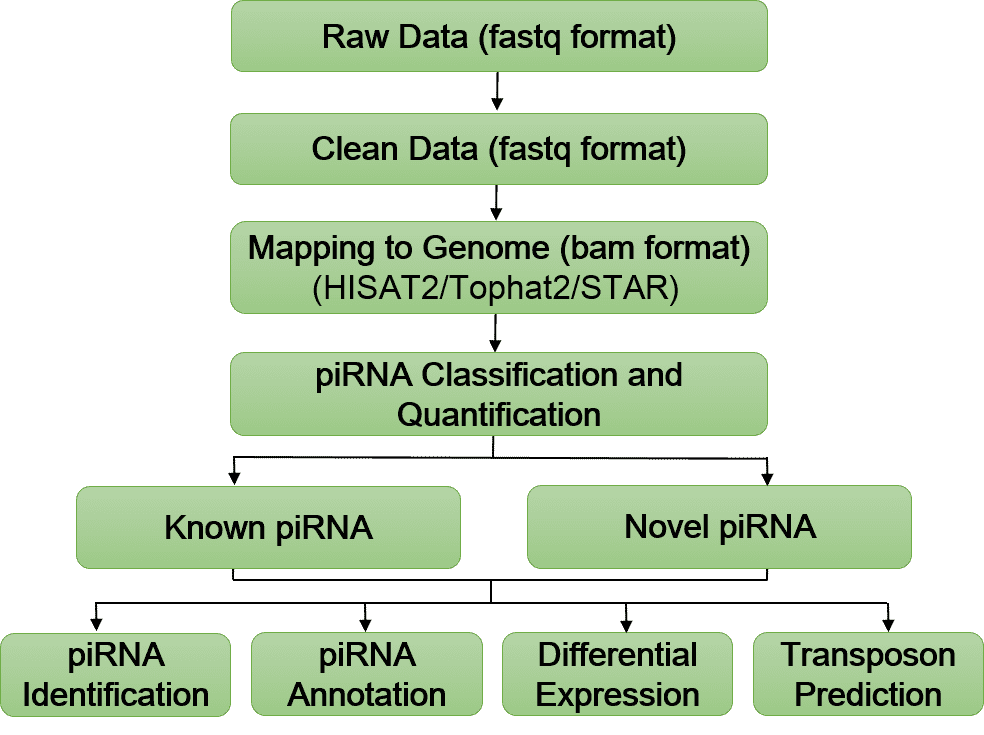
Bioinformatics Analysis Pipeline
In-depth data analysis:
- Genome-wide distribution of piRNA expression
- Differential expression analysis of piRNA
- Prediction and annotation of transposon element
- Identify known piRNA and predict new piRNA
- Cluster analysis
- Novel piRNA prediction
- piRNA target prediction
- Genome-wide ORF and CDS annotation
Sample Requirements
RNA sample quantity ≥ 2 ug, OD260/280 ≥ 1.8, OD260/230 ≥ 1.5.
Please make sure that the RNA is not significantly degraded.
Sample storage: The RNA sample can be dissolved in ethanol or RNA-free ultra-pure water and stored at -80°C. Sample should avoid repeated freezing and thawing.
Shipping Method: The RNA sample is stored in 1.5 mL Eppendorf tube, sealed with sealing membrane and transported with dry ice.
Deliverable: FastQ, BAM, coverage summary, QC report, custom bioinformatics analysis.
References:
- Perera B P U, Tsai Z T, Colwell M L, et al. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics, 2019, 5, 14(5): 504-521.
- Yang Q, Li R, Lyu Q, et al. Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat Commun, 2019, 7, 10(1): 3389-3396.
- Fabio M, Dominik H, Julius Brennecke. piRNA-guided slicing specifies transcripts for Zucchini dependent, phased piRNA biogenesis. Science, 2015, 348: 812-817.

