Long RNA-seq (lncRNA+circRNA+mRNA)
Long RNA sequencing (RNA-seq) is a next-generation sequencing (NGS)-based method to comprehensively detect long RNA sequences (>200 nt), including message RNA and non-coding RNA (ncRNA). It can accurately reveal the complete picture of the transcriptome in a biological sample under specific conditions. We provide long RNA-seq to help you analyze mRNA, long non-coding RNA (lncRNA), and circular RNA (circRNA) at one time.
Overview
RNA is a single-stranded ribonucleic acid molecule transcribed from DNA. It has many critical roles in coding, decoding, regulation and expression of genes. Cellular organisms use mRNA to convey genetic information that directs the synthesis of specific proteins. In addition to mRNA, there are many types of non-coding RNAs (ncRNAs) that cannot be translated into protein. They function at the RNA level, not protein level. LncRNAs and circRNAs are two types of common long ncRNAs. LncRNAs are defined as being non-coding transcripts with lengths exceeding 200 nt. Many lncRNAs are found to be functionally associated with human diseases, in particular, cancers. CircRNAs are covalently closed continuous loops, generally formed by alternative splicing of pre-mRNA. Evidences are arising that cirRNAs can regulate microRNA function and play a vital role in cancer-related or non-cancer pathways. RNA-seq is a technique that examines and quantity and sequences of RNA molecules in a sample using NGS. Long RNA-seq is specifically designed to obtain information on long RNA molecules exceeding 200 nt. Long RNA-seq experiments contribute to our understanding of novel transcripts and transcript variants, and how RNA molecules affect gene regulation, and thus disease development and phenotypic variation. We also provide tumor long RNA-seq and exosome long RNA-seq for medical research.
Features
| Any Species | Transcriptome-Wide | Bioinformatics Analysis | One-Stop Solution |
|---|---|---|---|
| This method can be applied to any species, from microorganisms to humans. | Profile all lncRNAs, circRNAs, mRNAs, either known or unknown, in your biological sample. | Our integrated bioinformatics pipeline can be tailored to suit your project. | One-stop solution from sample QC, library construction, to sequencing and data analysis. |
Project Workflow

1. Sample Preparation
Quality assessment and quantification; ribosomal RNA depletion and fragmentation

2. Library Preparation
Strand-specific libraries are constructed, and assessed and quantified by BioAnalyzer and RTQ-PCR

3. Sequencing
HiSeq X10/4000; PE50/75/100/150;
>10G clean data

4. Data Analysis
Visualize and preprocess results, and perform custom bioinformatics analysis
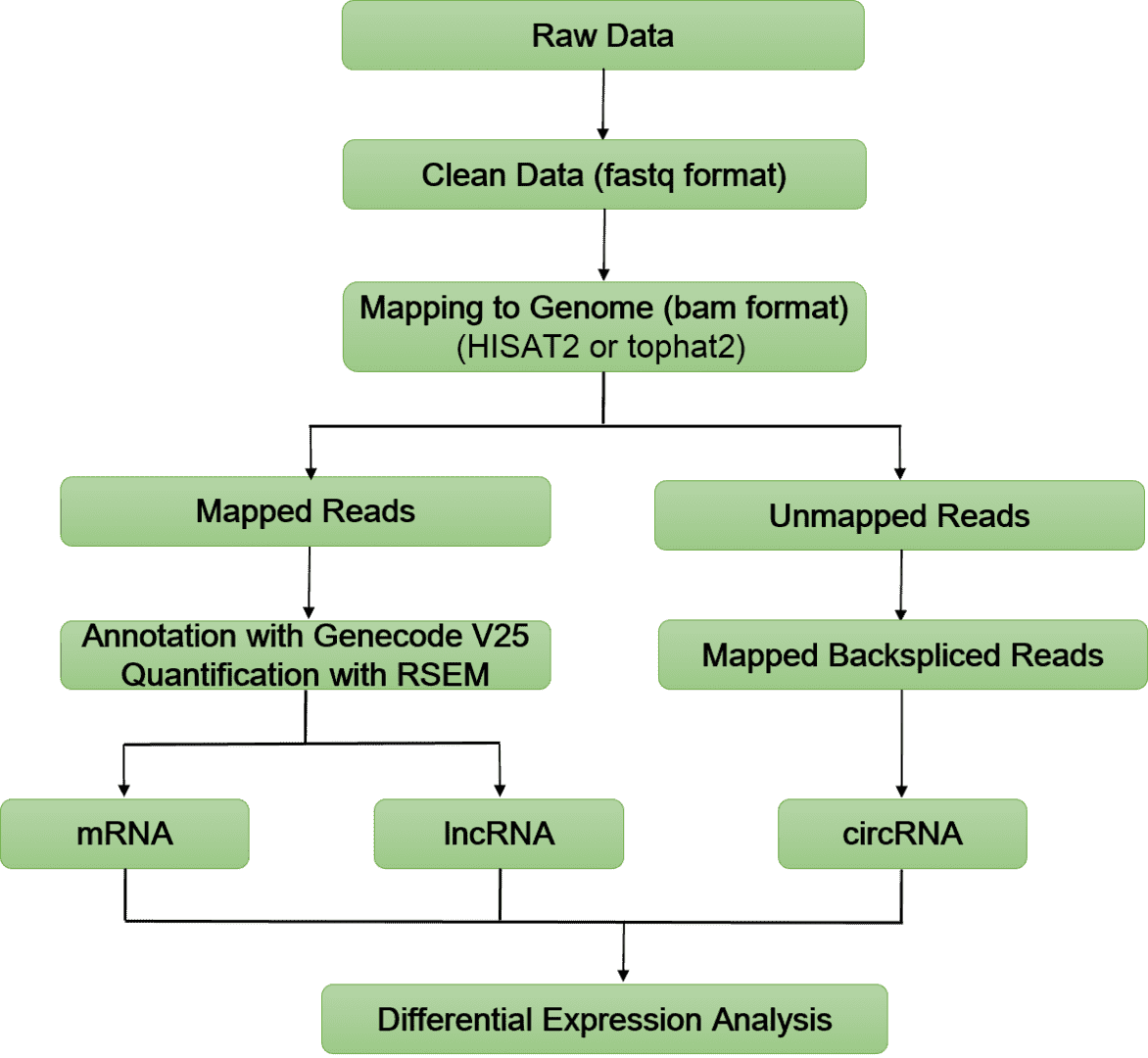
Bioinformatics Analysis Pipeline
In-depth data analysis:
- Profiling of lncRNA, circRNA and mRNA
- Predict novel lncRNA, circRNA and mRNA
- Detect novel and rare transcript variants
- Differential expression analysis of transcripts
- Target gene prediction and functional analysis of lncRNAs and circRNA
- GO analysis
- KEGG analysis
- Annotation of circRNA, lncRNA
Sample Requirements
- RNA sample (concentration ≥ 200 ng/uL, quantity ≥ 4 ug)
- 1.8 ≤ OD260/280 ≤ 2.2, OD260/230≥2.0, RIN ≥ 6.5, 28S:18S≥1.0.
- Please make sure that RNA is not degraded.
Sample storage: RNA can be dissolved in ethanol or RNA-free ultra-pure water and stored at -80℃. RNA should avoid repeated freezing and thawing.
Shipping Method: When shipping RNA samples, the RNA sample is stored in a 1.5 mL Eppendorf tube, sealed with a sealing film. Shipments are generally recommended to contain 5-10 pounds of dry ice per 24 hours.
Deliverable: FastQ, BAM, coverage summary, QC report, custom bioinformatics analysis.






