MeRIP Sequencing (m6A-seq)
Methylated RNA immunoprecipitation sequencing (MeRIP-seq) is a next-generation sequencing (NGS)-based method to comprehensively detect m6A. RNA methylation modification exists in various transcripts. We provide one-stop MeRIP-seq services to meet various in-depth data analysis needs of customers.
Overview
Various post-transcriptional modifications are present in eukaryotes mRNA. N6-methyladenosine (m6A) is the most common and abundant modification in the transcriptome. m6A is also highly prevalent in mRNAs as well as in certain non-coding RNAs. m6A is typically near the stop codon, but also in the coding sequence, 3' UTR, and 5' UTR of mRNAs. Studies have shown that m6A modification has multiple effects on mRNA fate - it regulates different stages of mRNA metabolism, including folding, maturation, export, translation, and decay, and further promotes many biological processes, including circadian rhythm, adipogenesis, and embryonic stem cell renewal. The importance of m6A modification has become a hot spot for epigenetic transcriptome research. MeRIP-seq combines methylated DNA immunoprecipitation technology, RNA-binding protein immunoprecipitation technology, and RNA sequencing technology to detect the complete map of RNA methylation with high precision. The technique uses immunoprecipitation to co-incubate methylated RNA-specific antibodies with randomly interrupted RNA fragments, and grabs methylated modified fragments for sequencing. In the meantime, the control sample needs to be sequenced in parallel. Later, sequence fragments in the co-immunoprecipitation sample and control sample are mapping to the reference genome/ transcriptome to detect RNA methylation sites. As a mainstream method for detecting m6A modifications, MeRIP-seq can comprehensively analyze the distribution and changes of m6A in the transcriptome, helping researchers deeply understand the role of RNA modifications in physiology and pathology, thus, promoting accurate diagnosis and treatment of diseases.
Features
| Any Species | High Resolution | High Accuracy | Digitized Signal |
|---|---|---|---|
| This method can be applied to any species, from microorganisms to humans. | Accurate peak positioning within 100 bases of the m6A sites. | m6A levels calibrated with the transcript abundance in the input control. | Direct quantification and sequencing of methylated fragments without cross-reactivity and background noise. |
Project Workflow

1. Sample Preparation
RNA purification;
quality assessment and quantification.

2. Library Preparation
Poly(A) tail RNA selection;
m6A immunoprecipitation;
cDNA library preparation.

3. Sequencing
Illumina HiSeq;
PE 50/75/100/150.

4. Data Analysis
Visualize and preprocess results, and perform custom bioinformatics analysis.
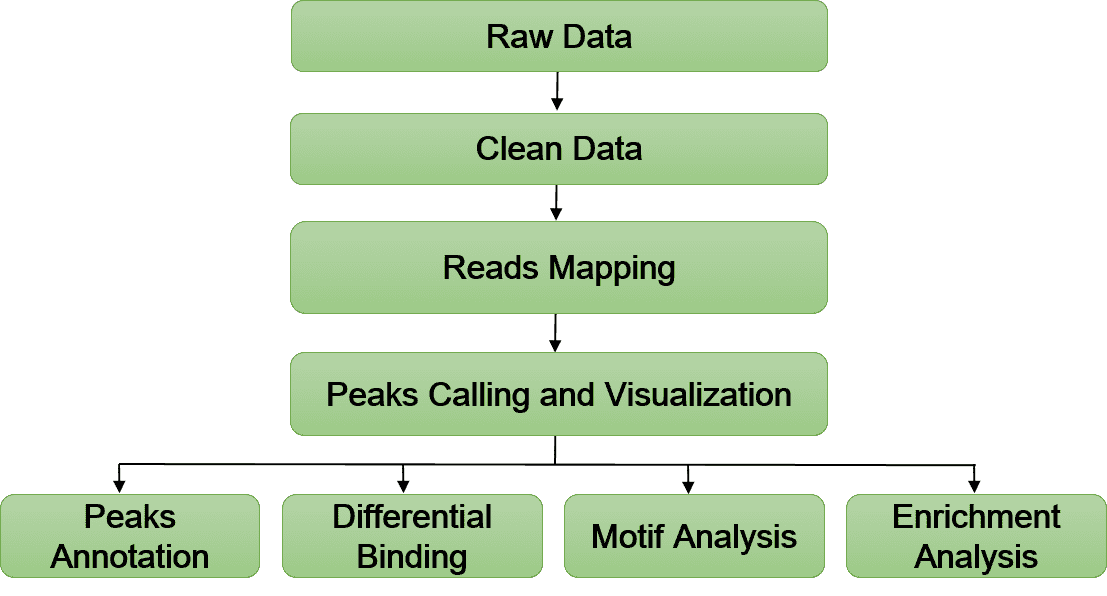
Bioinformatics Analysis Pipeline
In-depth data analysis:
- Biostatical analysis of m6A data
- Peaks annotation
- Transcriptome-wide profiling of m6A methylation
- Differential methylation
- Differential peak analysis
- Motif identification of enrichment sites
- GO and KEGG pathway analysis
Sample Requirements
RNA sample quantity ≥ 50 ug.
OD260/280 ≥ 1.8, OD260/230 ≥ 1.0, RIN ≥ 7.0.
Please make sure that the RNA is not significantly degraded.
Sample storage: RNA can be dissolved in ethanol or RNA-free ultra-pure water and stored at -80°C. RNA should avoid repeated freezing and thawing.
Shipping Method: When shipping RNA samples, the RNA sample is stored in a 1.5 mL Eppendorf tube, sealed with sealing film. Shipments are generally recommended to contain 5-10 pounds of dry ice per 24 hours.
Deliverable: FastQ, BAM, coverage summary, QC report, custom bioinformatics analysis.
References:
- Meyer K D, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell, 2012, 149(7):1635-1646.
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 2012, 485(7397): 201-206.
- Clancy M J, Shambaugh M E, Timpte C S, et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucl Acids Res, 2002, 30(20): 4509-4518.




