2'-O-Methylation Sequencing
2'-O-Methylation sequencing is a next generation sequencing (NGS)-based method to comprehensively detect 2'-O-RNA modification at single-base resolution and of high detection sensitivity. It can quickly identify the complete picture of the 2'-O-RNA methylation modification in transcripts. We provide one-stop 2'-O-Methylation sequencing service to meet various in-depth data analysis needs of customers.
Overview
2'-O-methylation modification is a chemical modification in which ribosomal RNA is methylated at 2' position under the action of RNA methylase or fibrinase. Studies have shown that 2'-O-methylation modification is mainly present in specific residues of transfer RNA, ribosomal RNA, small nuclear RNA, and 5'-cap of mammalian cell mRNA. It can affect the combination of mRNA and protein, regulate the efficiency of rRNA translation, and participate in innate immunity and biological processes such as tRNA recognition. Combining high-throughput sequencing technology with bioinformatics analysis methods, our company provides one-stop 2'-O-methylation sequencing service and detailed data reports, which can systematically analyze 2'-O-methylation modification within the whole transcriptome. The feature is that sodium periodate can specifically oxidize the sites without 2'-O-methylation, after multiple cycles of purification, the proportion of methylated fragments in sequencing library can be greatly increased. 2'-O-methylation sequencing is suitable for multiple molecules, including mRNA, lncRNA, pri-miRNA, tRNA and rRNA. 2'-O-methylation sequencing assays contribute to our understanding of transcript variants and how RNA molecules affect gene regulation that associates with disease development and phenotypic variation.
Features
| Single-base Resolution | Wide Detection Range | High Coverage | Bioinformatics Service |
|---|---|---|---|
| Single-base resolution allows accurate detection of 2'-O-RNA methylation modification. | Detection range can cover all methylated regions in transcriptome. | Detect 2'-O-RNA methylation sites of multiple RNA molecules, including mRNA, lncRNA, pri-miRNA, tRNA, rRNA, etc. | Provide standard and customized bioinformatics analyses, which are reflected in the report. |
Project Workflow

1. Sample Preparation
RNA purification;
quality assessment and quantification.

2. Library Preparation
RNA fragmentation;
specific library preparation.

3. Sequencing
Illumina HiSeq;
PE 50/75/100/150.

4. Data Analysis
Visualize and preprocess results, and perform custom bioinformatics analysis.
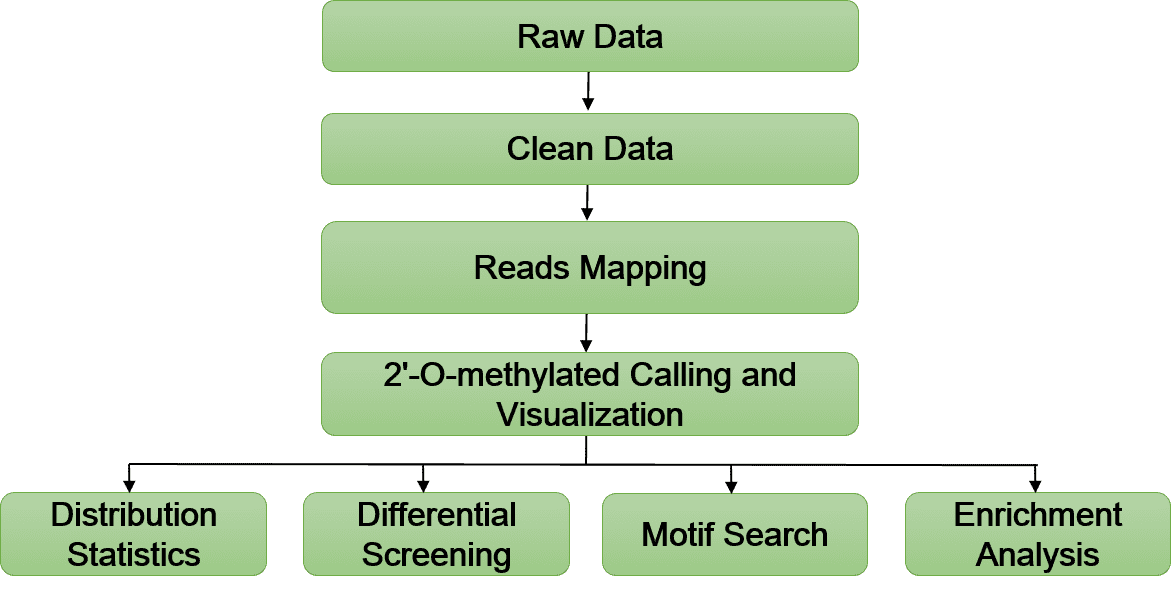
Bioinformatics Analysis Pipeline
In-depth data analysis:
- Statistics of 2'-O-RNA signal distribution
- 2'-O-RNA methylated transcriptome map analysis
- 2'-O-RNA methylated calling
- Evolutionary conservative analysis
- Motif search
- Differential gene screening of enrichment sites
- KEGG pathway enrichment analysis
- Gene ontology enrichment analysis
Sample Requirements
RNA sample quantity ≥ 100 ug.
1.6 ≤ OD260/280 ≤ 2.3.
Please make sure that the RNA is not significantly degraded.
Sample storage: RNA can be dissolved in ethanol or RNA-free ultra-pure water and stored at -80°C. RNA should avoid repeated freezing and thawing.
Shipping Method: When shipping RNA samples, the RNA sample is stored in a 1.5 mL Eppendorf tube, sealed with sealing film. Shipments are generally recommended to contain 5-10 pounds of dry ice per 24 hours.
Deliverable: FastQ, BAM, coverage summary, QC report, custom bioinformatics analysis.
References:
- Dai Q, Moshitch-Moshkovitz S, Han D, et al. Nm-seq maps 2'-O-methylation sites in human mRNA with base precision. Nature Methods, 2017, 15, 14(7): 695-698.
- Nicolai K, Martin D J, Sophia J H, et al. Profiling of 2'-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Research, 2016, 16, 19: 7884-7895.
- Yin Z Z, Stephan P P, Gordon G C. High-throughput and site-specific identification of 2'-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA, 2017, 8, 23(8): 1303-1314.
- Yuri M, Virginie M. Detection and Analysis of RNA Ribose 2'-O-Methylations: Challenges and Solutions. Genes (Basel), 2018, 12, 9(12): 642.



